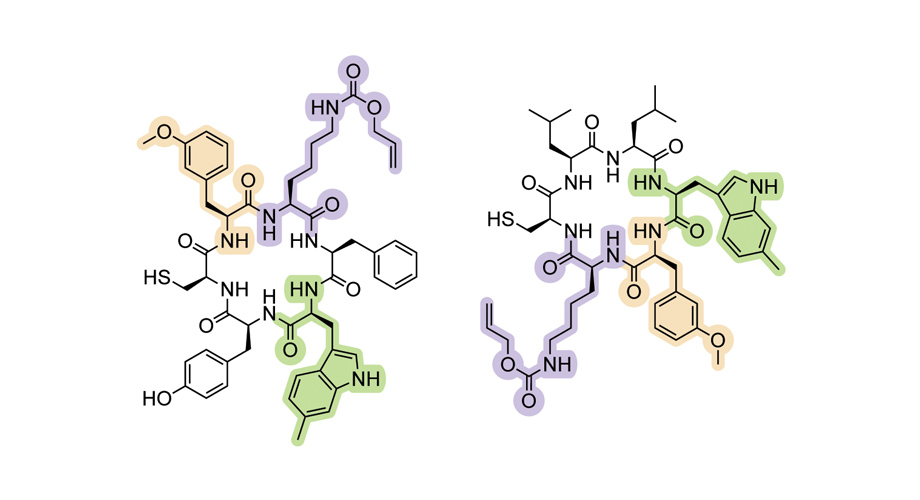
Researchers at Scripps Research have created a new paradigm that expands the genetic alphabet as we know it by engineering biologic molecules. As a typical protein is built from 20 amino acids encoded in mRNA by triplet codons, the new method done at Scripps use four RNA nucleotides instead of three. This new technique will allow for easy addition of non-canonical amino acids to proteins.
The engineering of unique transfer RNAs (tRNAs) correspond with the four-letter codons shows that editing one gene will incorporate the synthetic amino acids by using the cell's ability to synthesize proteins.Using this successful technique allowed scientists to generate over 100 new cyclic peptides (aka macrocycles), where all contain no more than three non-canonical amino acids. This result bypasses the need to rewrite the organism's entire genome, allowing for a flexible, efficient application to tailoring proteins.
Using this successful technique allowed scientists to generate over 100 new cyclic peptides (aka macrocycles), where all contain no more than three non-canonical amino acids. Lead researcher Dr. Ahmed Badran exclaims "our results suggest that one can now easily and effectively incorporate non-canonical amino acids at diverse sites in a wide array of proteins."
This newfound ability, when perfected, will broaden the horizons for future biological endeavors and presently, more research will only benefit the scientific community as a whole.
Sources:
https://www.scripps.edu/news-and-events/press-room/2024/2024911-badran-rna-nucleotides.html
https://pubs.acs.org/doi/10.1021/acs.chemrev.5c00065
* sorry for black/ white highlighting, formatting was affected when taking quotes from study
This is a really fascinating breakthrough — expanding the genetic alphabet feels like something straight out of synthetic biology textbooks, but seeing it actually work in living cells is incredible. I like how you highlighted that the team didn’t need to rewrite an entire genome to incorporate these non-canonical amino acids. That’s a major advantage because it keeps the system flexible and avoids the massive challenges of building an organism with fully redesigned DNA.
ReplyDeleteWhat stood out to me most was the ability to use four-letter codons. That dramatically increases the coding capacity and opens the door to designing proteins with properties nature never produced on its own. The fact that they generated over 100 new macrocycles shows just how powerful this method already is.
Overall, this type of work could change how we approach drug development, enzyme engineering, and even biomaterials. It definitely feels like a big step forward for synthetic biology and protein design. Great post — very interesting topic!
That is an amazing advancement. So much can be done with this -- the sky's the limit. I really hope that this gets studied more, so that it can be used more often and more effectively.
ReplyDelete