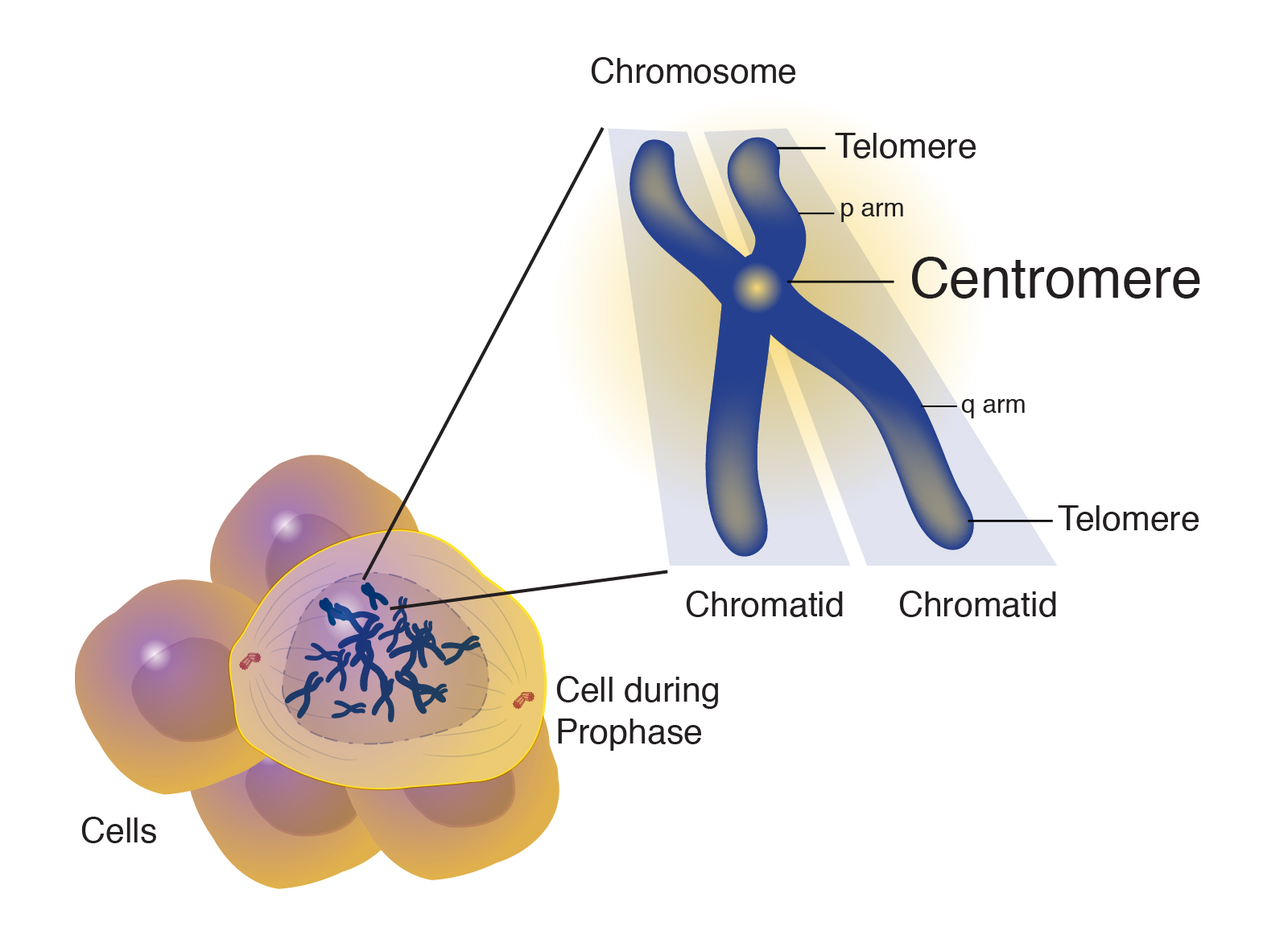
The researchers first looked for single nucleotide polymorphisms in Drosophila, which allowed them to map haplotypes. Langley stated that this finding had two implications, Firstly, if researchers can distinguish chromosomes from each other by their centromeres, they can start to carry out functional tests to see if these differences have an impact on which piece of DNA is inherited. This could allow researches to determine if a single centromere haplotype is transmitted more often or if a certain haplotype is more prone to errors. Secondly, the centromeres could be analyzed to determine evolutionary descent. In the 1000 Genomes Project, these haplotypes were also found spanning over human centromeres.
Several haplotypes were found within the centromeres of human X chromosomes. In these DNA analyses, most of the diversity is seen among African genomes. Chromosome 11 showed highly diverged stretches of neanderthal DNA in non-african genomes. Humans have roughly 400 different genes for odorant receptors, 34 of which are found within the centromere of chromosome 11. It was found that Neanderthal haplotypes within centromeres account for roughly half of the variation in odorant receptors which influence taste and smell. This research has the potential to further our knowledge regarding human evolution in determining specific ancestral patterns of large groups of people.
https://www.sciencedaily.com/releases/2019/06/190618105024.htm
No comments:
Post a Comment